The ChemPert web interface mainly includes two sections: the database and the webtool, which reveals the relationship between external perturbations, protein targets of perturbations and downstream transcriptional signatures in non-cancer cells.
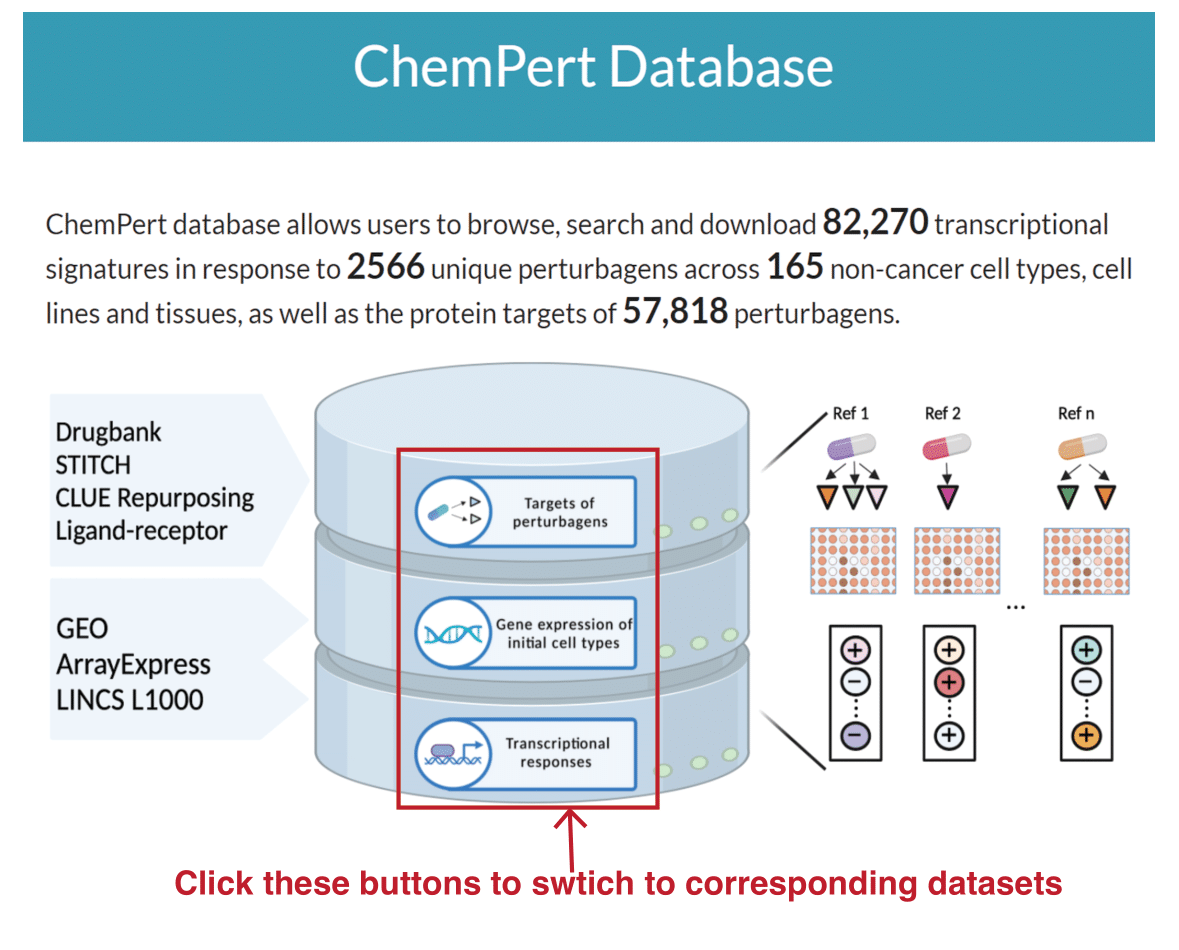
The home page of the database provides a summary of the database and allows users to get access to one of the three main resources of the databases, the targets of perturbagens, the gene expression profiles of initial cellular states and the TF responses after perturbations. To browse, search and download the resource of the ChemPert database, please click the corresponding buttons:
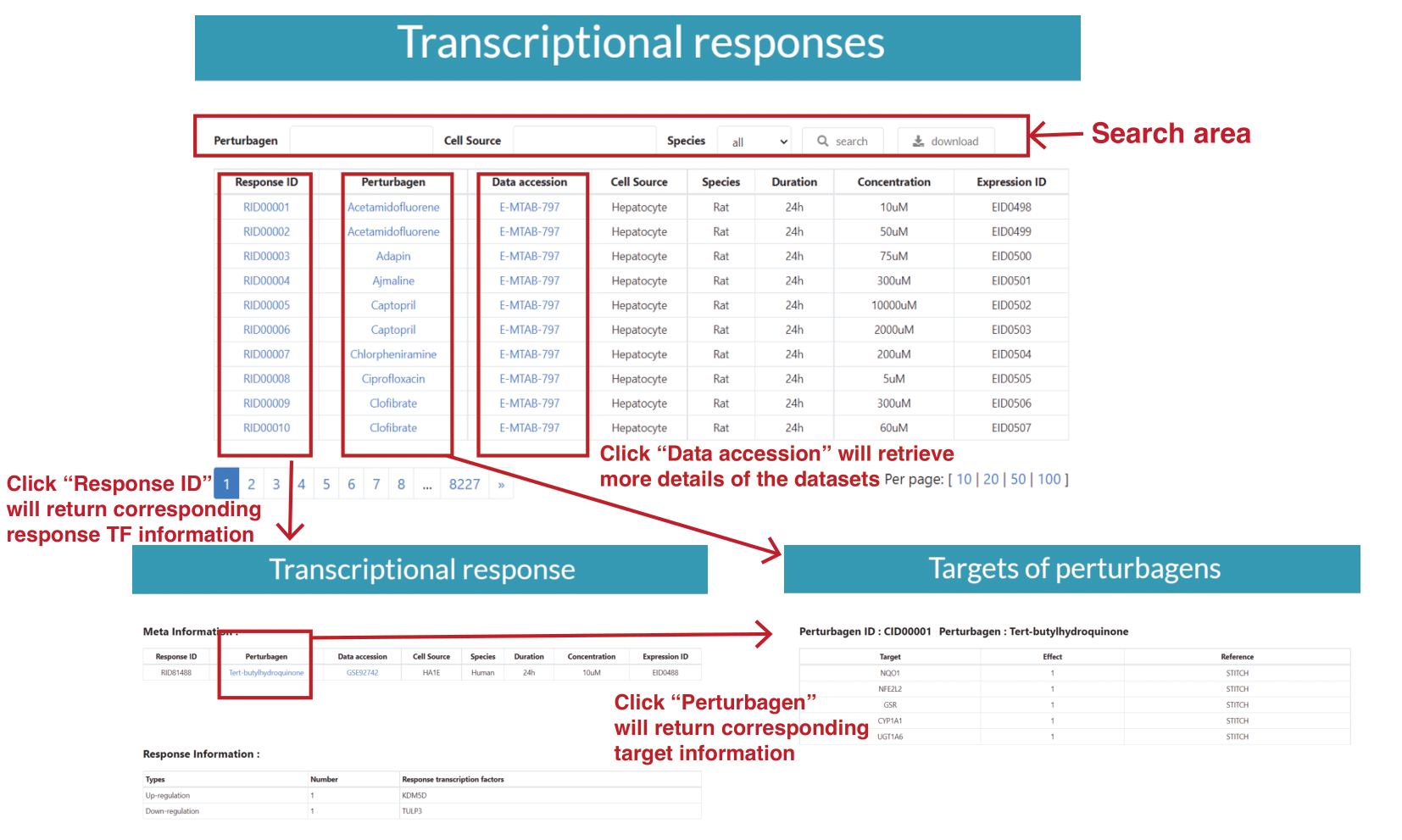
For example, when users click the button “Transcriptional responses”, a table listing the major meta information on each dataset will be returned, including the perturbagen, data accession number, cell type, perturbation duration and concentration. The search area allows users to search for the datasets of interest based on the perturbagens, cell types or species. Then users can download or browse the datasets of interest. In particular, users can click the “Response ID” to browse the response TFs of corresponding dataset. Clicking the “Perturbagen” button enables the users to browse the protein targets of this perturbagen. To retrieve the details of original dataset, users can click “Data accession”.
To achieve the batch download of the whole database, users can go to Download page.
The webtool section provides an intuitive interface for users to predict either response TFs or perturbagens.
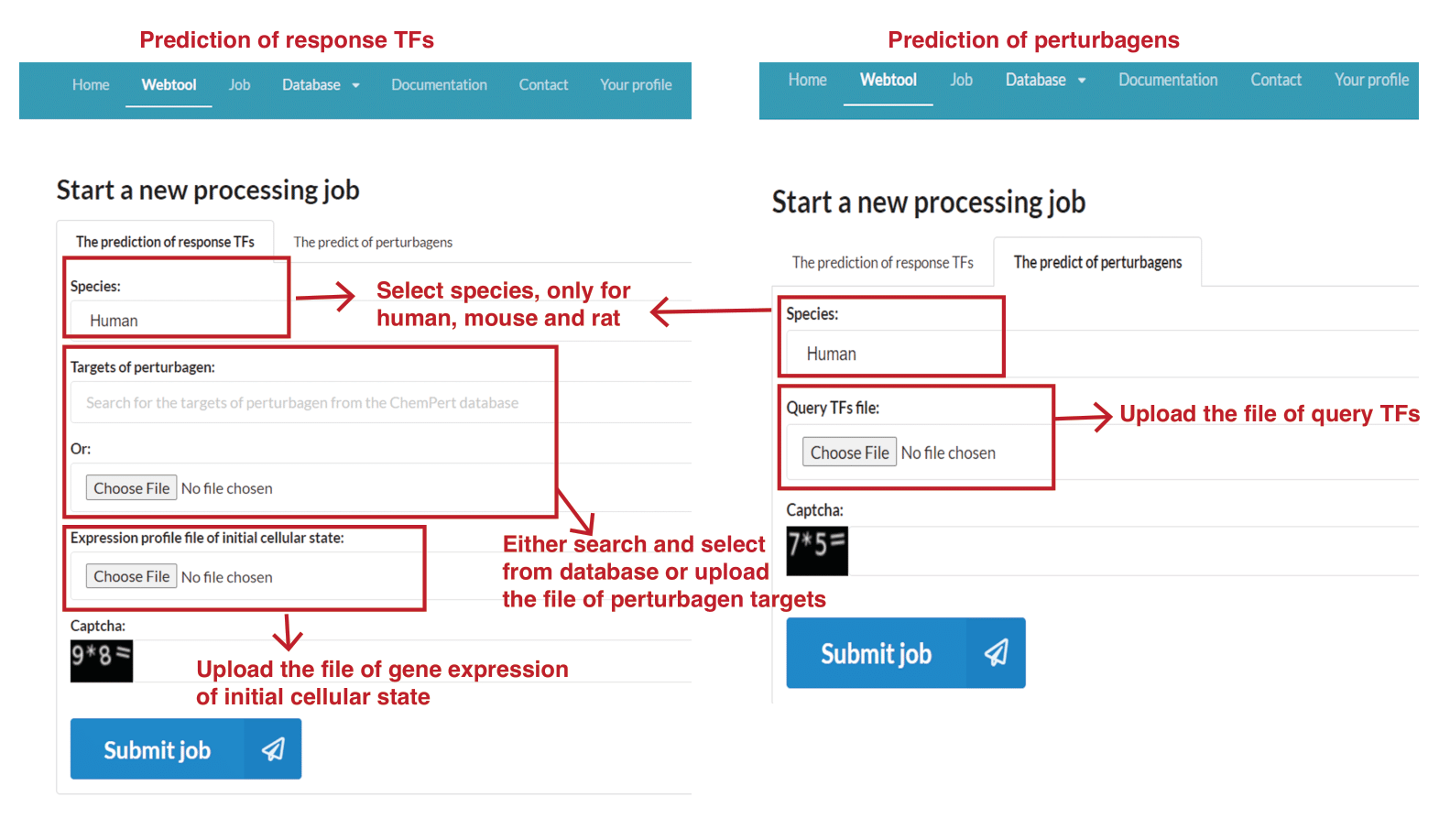
1. Go to "Webtool" subpage, set the parameters and submit the input files. The webtool allows users to predict either response TFs or perturbagens.
For the prediction of response TFs, users need to set the following three parameters (see the following figure):
a) Species: human/mouse/rat.
b) Targets of perturbagen. Users can search for the targets of perturbagen in the ChemPert database as input. If a query perturbagen is not available in the database, users can upload the protein targets of the query perturbagen.
c) Gene expression profile of initial cellular state. Initial expression file must have gene symbol as rownames and one column with mean expression level of corresponding gene.
The webtool accepts the input files of perturbagen targets and gene expression profile in either tab-delimited text file (.txt file) or tab-delimited R object file (.rds file). Users can check the example of the input and output files in Download page.
For the prediction of perturbagens, users need to set the following two parameters (see the following figure):
a) Species: human/mouse/rat.
b) Query TFs
The webtool accepts the input file of query TFs in either tab-delimited text file (.txt file) or tab-delimited R object file (.rds file). Users can check the example of the input and output files in Download page.
2. Wait for the processing to finish (it might take up to a couple of hours for the prediction of response TFs).
If you didn't submit your e-mail address, do not close the page - but refresh it from time to time to track the progress.
3. If you submitted your e-mail address, you will be notified by e-mail when the processing finishes. Users can track the status of the job and download the final predictions in “Jobs” page.
Output files for the prediction of response TFs:
a) padjust_enriched_allSimplePath_MajorityLen.Robj: This file contains the p-adjusted value of the enriched short paths for the initial gene expression data. This file is used for the prediction of response TFs.
b) predicted_reTFs.txt: This file contains the list of predicted response TFs that are sorted by the frequency with which each TF appeared in retrieved transcriptomics datasets. The higher this frequency, the more likely that the TF is a responder of the query perturbation.
Note: if the query initial state is not similar enough with reference data in the database, predicted_reTFs.txt file will not be generated and there is note “No reference data was retrieved because the query initial cell state is not similar enough” in log file.
Output files for the prediction of perturbagens:
a) predicted_signalling_proteins.xlsx: This file contains the list of predicted signalling proteins.
b) predicted_perturbagens.xlsx: This file contains the list of predicted perturbagens and their corresponding information.
Due to the limited number of CPUs available for this web application, your jobs may be in the waiting queue for long time, but they will eventually be processed. If the waiting time is too long, please contact us.